
Introduction to Pandas
In the previous notebook, we dove into the details on NumPy, which provides efficient storage and ability to perform complex computations through its ndarray object for working with homogeneous numerical array data. Here, we will build on the knowledge by looking into the data structures provided by Pandas.
Pandas provides high-level data structures and functions designed to make working with structured or tabular data fast, easy, and expressive. It is based on two main data structures:
Series: one-dimensional such as a list of itemsDataFrame: two-dimensional, such as a table
Both Series and DataFrame objects build on the NumPy array structure and form the core data model for Pandas in Python. While NumPy is best suited for working with homogeneous data, Pandas is designed for working with tabular or heterogeneous data. NumPy serves its purpose very well when the data is well organized and clean, however its limitations become clear when working with less structured data where we need more flexibility such as attaching labels to data, working with missing data, grouping, pivoting etc.
Pandas blends the high-performance, array-computing ideas of NumPy with the flexible data manipulation capabilities of relational databases (such as SQL). Pandas provides sophisticated indexing functionality to reshape, slice and dice, perform aggregations, and select subsets of data. It also provides capabilities for easily handling missing data, adding/deleting columns, imputing missing data, and creating plots on the go. Pandas is a must-have tool for data wrangling and manipulation.
Installation and Import
A typical installation of Python API comes with Pandas. You can import it as:
import pandas as pd
import numpy as npYou can also check the version of Pandas that is installed:
pd.__version__'1.0.5'
Working with Series
A Pandas Series is a one-dimensional array of indexed data. A Series object contains a sequence of values and an associated array of data labels, called index. While Numpy Array has an implicitly defined integer index that can be used to access the values, the index for a Pandas Series can also be explicitly defined. Let's take a look at some examples.
Creating Series from a List or Array
A Series can be created from a list or array as follows:
# Create Series
data1 = [1,2,3,6,7]
s1 = pd.Series(data1)
s10 1 1 2 2 3 3 6 4 7 dtype: int64
The array representation and index object of the Series can be accessed via its values and index attributes:
print('Values in series s1:', s1.values)
print('Index of series s1:', s1.index)Values in series s1: [1 2 3 6 7] Index of series s1: RangeIndex(start=0, stop=5, step=1)
Like NumPy arrays, data in a Series can be accessed by the associated index.
# Access elements of a Series
print(s1[0])
print(s1[3])1 6
For Series objects, the index need not be an integer and can be explicitly defined as follows:
s2 = pd.Series([1,2,3,6,7], index=['d', 'b', 'a', 'c', 'e'])
s2d 1 b 2 a 3 c 6 e 7 dtype: int64
# Check the index
s2.indexIndex(['d', 'b', 'a', 'c', 'e'], dtype='object')
# Access elements using index label
print('Element associated with index a:', s2['a'])
print('Element associated with index e:', s2['e'])Element associated with index a: 3 Element associated with index e: 7
# Access elements using index position
print('Element associated with index position 2:', s2[2])
print('Element associated with last index position:', s2[-1])Element associated with index position 2: 3 Element associated with last index position: 7
Creating Series from a Dictionary
Just like a dictionary maps keys to a set of values, a series can be thought of as a mapping of index values to data values. Let's create a series from a dictionary.
data = {'CA': 35000, 'TX': 71000, 'WA': 16000, 'OR': 5000}
s3 = pd.Series(data)
s3CA 35000 TX 71000 WA 16000 OR 5000 dtype: int64
By default, index for Series will be created using the keys.
# Access elements using index label
s3['CA']35000
# Access elements using index position
s3[0]35000
Working with DataFrame
A DataFrame represents a rectangular table of data and contains an ordered collection of columns. You can think of it as a spreadsheet or SQL table where each column has a column name for reference and each row can be accessed by using row numbers. Column names and row numbers are known as column and row index.
DataFrame is a fundamental Pandas data structure in which each column can be of a different value type (numeric, string, boolean, etc.). A data set can be first read into a DataFrame and then various operations (i.e. indexing, grouping, aggregation etc.) can be easily applied to it.
Creating a DataFrame
There are many ways to construct a DataFrame. Let's look at some examples.
From a Dictionary
DataFrame can be constructed from a dictionary of equal sized lists or NumPy arrays.
data = {'state':['CA','WA','CA','WA','CA','WA'],
'year':[2015,2015,2016,2016,2017,2017],
'population':[38.5,7.0,39.0,7.25,39.5,7.5]}
df1 = pd.DataFrame(data)
df1| state | year | population | |
|---|---|---|---|
| 0 | CA | 2015 | 38.50 |
| 1 | WA | 2015 | 7.00 |
| 2 | CA | 2016 | 39.00 |
| 3 | WA | 2016 | 7.25 |
| 4 | CA | 2017 | 39.50 |
| 5 | WA | 2017 | 7.50 |
From a Series
DataFrame is a collection of Series objects. Let's look at constructing a DataFrame from a single Series object.
# Create Population series
pop_data = {'CA': 39.5,'TX': 29,'NY': 8.39,'FL': 21.48,
'IL': 12.67}
population = pd.Series(pop_data)# Create DataFrame from series
pop_df = pd.DataFrame(population, columns=['population'])
pop_df| population | |
|---|---|
| CA | 39.50 |
| TX | 29.00 |
| NY | 8.39 |
| FL | 21.48 |
| IL | 12.67 |
From a Dictionary of Series
DataFrame can also be constructed from a dictionary of Series.
# Create Area series
area_data = {'CA': 155779.22, 'TX': 261231.71, 'NY': 47126.40,
'FL': 53624.76, 'IL': 55518.93}
area = pd.Series(area_data)
areaCA 155779.22 TX 261231.71 NY 47126.40 FL 53624.76 IL 55518.93 dtype: float64
# Create DataFrame from dictionary of Series
df2 = pd.DataFrame({'population':population,
'land area':area})
df2| population | land area | |
|---|---|---|
| CA | 39.50 | 155779.22 |
| TX | 29.00 | 261231.71 |
| NY | 8.39 | 47126.40 |
| FL | 21.48 | 53624.76 |
| IL | 12.67 | 55518.93 |
From a two-dimensional NumPy Array
DataFrame can be constructed from a two-dimensional NumPy array by specifying the column names. An integer Index is used if not specified.
pd.DataFrame(np.random.rand(4, 2),
columns=['Col_A', 'Col_B'],
index=['a', 'b', 'c', 'd'])| Col_A | Col_B | |
|---|---|---|
| a | 0.028278 | 0.687096 |
| b | 0.999634 | 0.443590 |
| c | 0.750714 | 0.785287 |
| d | 0.611652 | 0.651515 |
Importing Data into a DataFrame
Reading data into a DataFrame is one of the most common task in any data science problem. Pandas provides the ability to read data from various formats such as CSV, JSON, Excel, APIs, etc. directly into a DataFrame object. Let's look at how to read data from some common formats into a DataFrame.
Read from a csv
read_csv() function can be used to read csv (comma-separated value) files. The function includes a number of different parameters and you can read more about them in the pandas documentation here.
Let's import the 'health.csv' file we used earlier in this guide series.
# Read the data from csv
df_csv = pd.read_csv('../data/health.csv')
df_csv.head()| Number of Beds | Name | Address | City | State | Zip Code | |
|---|---|---|---|---|---|---|
| 0 | 156 | Facility 1 | 2468 SOUTH ST ANDREWS PLACE | LOS ANGELES | CA | 90018 |
| 1 | 59 | Facility 2 | 2300 W. WASHINGTON BLVD. | LOS ANGELES | CA | 90018 |
| 2 | 25 | Facility 3 | 4060 E. WHITTIER BLVD. | LOS ANGELES | CA | 90023 |
| 3 | 49 | Facility 4 | 6070 W. PICO BOULEVARD | LOS ANGELES | CA | 90035 |
| 4 | 55 | Facility 5 | 1480 S. LA CIENEGA BL | LOS ANGELES | CA | 90035 |
# Confirm type
type(df_csv)pandas.core.frame.DataFrame
Read from an excel file
Tabular data is often stored using Microsoft Excel 2003 (and higher) and can be read using read_excel().
# Read the data from excel
df_excel = pd.read_excel('../data/health.xlsx')
df_excel| Number of Beds | Name | Address | City | State | Zip Code | |
|---|---|---|---|---|---|---|
| 0 | 156 | Facility 1 | 2468 SOUTH ST ANDREWS PLACE | LOS ANGELES | CA | 90018 |
| 1 | 59 | Facility 2 | 2300 W. WASHINGTON BLVD. | LOS ANGELES | CA | 90018 |
| 2 | 25 | Facility 3 | 4060 E. WHITTIER BLVD. | LOS ANGELES | CA | 90023 |
| 3 | 49 | Facility 4 | 6070 W. PICO BOULEVARD | LOS ANGELES | CA | 90035 |
| 4 | 55 | Facility 5 | 1480 S. LA CIENEGA BL | LOS ANGELES | CA | 90035 |
# Confirm type
type(df_excel)pandas.core.frame.DataFrame
Read from a json
read_json() can be used to read JSON (JavaScript Object Notation) files. JSON is mostly used to store unstructured data with key/value pairs. The function accepts a valid JSON string, path object or file-like object and does not consume a dictionary (key/value pair) directly. Let's read 'health.json' into a DataFrame.
# Read the data from json
df_json = pd.read_json('../data/health.json')
df_json| Number of Beds | Name | Address | City | State | Zip Code | |
|---|---|---|---|---|---|---|
| 0 | 156 | Facility 1 | 2468 SOUTH ST ANDREWS PLACE | LOS ANGELES | CA | 90018 |
| 1 | 59 | Facility 2 | 2300 W. WASHINGTON BLVD. | LOS ANGELES | CA | 90018 |
| 2 | 25 | Facility 3 | 4060 E. WHITTIER BLVD. | LOS ANGELES | CA | 90023 |
| 3 | 49 | Facility 4 | 6070 W. PICO BOULEVARD | LOS ANGELES | CA | 90035 |
| 4 | 55 | Facility 5 | 1480 S. LA CIENEGA BL | LOS ANGELES | CA | 90035 |
# Confirm type
type(df_json)pandas.core.frame.DataFrame
Read from an API
Many websites provide data through public APIs in json or other formats. One easy way to access these APIs from Python is using requests package. Let's find issues for pandas on GitHub using the add-on requests library.
# Import library
import requests# Make request and store response
url = 'https://api.github.com/repos/pandas-dev/pandas/issues'
resp = requests.get(url)The response object’s json method will return a dictionary containing JSON. .keys() method can be used to explore the structure of the returned JSON object.
# Get json
json_data = resp.json()
json_data[0].keys()dict_keys(['url', 'repository_url', 'labels_url', 'comments_url', 'events_url', 'html_url', 'id', 'node_id', 'number', 'title', 'user', 'labels', 'state', 'locked', 'assignee', 'assignees', 'milestone', 'comments', 'created_at', 'updated_at', 'closed_at', 'author_association', 'active_lock_reason', 'pull_request', 'body', 'performed_via_github_app'])
Since each element in json_data is a dictionary, you can create a DataFrame using specific columns from the data.
# Create dataframe for specific cols
issues = pd.DataFrame(json_data, columns=['number', 'title',
'labels', 'state', 'created_at'])
issues.head()| number | title | labels | state | created_at | |
|---|---|---|---|---|---|
| 0 | 38070 | BUG: Index.drop raising Error when Index has d... | [] | open | 2020-11-25T20:11:11Z |
| 1 | 38069 | ENH: Rename multi-level columns or indices usi... | [{'id': 76812, 'node_id': 'MDU6TGFiZWw3NjgxMg=... | open | 2020-11-25T19:57:28Z |
| 2 | 38068 | ENH: NDArrayBackedExtensionArray.__array_funct... | [] | open | 2020-11-25T18:32:41Z |
| 3 | 38067 | DOC: Include missing holiday observance rules | [{'id': 134699, 'node_id': 'MDU6TGFiZWwxMzQ2OT... | open | 2020-11-25T17:25:35Z |
| 4 | 38066 | RLS: 1.1.5 | [{'id': 131473665, 'node_id': 'MDU6TGFiZWwxMzE... | open | 2020-11-25T15:51:33Z |
To create a DataFrame with all the data directly from the response object, "json" library can be used. Since read_json() accepts a valid JSON string, json.dumps() can be used to convert the object back to a string.
# Import library
import json# Read all data from response object's json method
all_issues = pd.read_json(json.dumps(json_data))
all_issues.head()| url | repository_url | labels_url | comments_url | events_url | html_url | id | node_id | number | title | ... | milestone | comments | created_at | updated_at | closed_at | author_association | active_lock_reason | pull_request | body | performed_via_github_app | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | https://api.github.com/repos/pandas-dev/pandas... | https://api.github.com/repos/pandas-dev/pandas | https://api.github.com/repos/pandas-dev/pandas... | https://api.github.com/repos/pandas-dev/pandas... | https://api.github.com/repos/pandas-dev/pandas... | https://github.com/pandas-dev/pandas/pull/38070 | 751090409 | MDExOlB1bGxSZXF1ZXN0NTI3NjcwMDk4 | 38070 | BUG: Index.drop raising Error when Index has d... | ... | None | 0 | 2020-11-25 20:11:11+00:00 | 2020-11-25 20:39:16+00:00 | NaT | MEMBER | NaN | {'url': 'https://api.github.com/repos/pandas-d... | - [x] closes #38051\r\n- [x] closes #33494\r\n... | NaN |
| 1 | https://api.github.com/repos/pandas-dev/pandas... | https://api.github.com/repos/pandas-dev/pandas | https://api.github.com/repos/pandas-dev/pandas... | https://api.github.com/repos/pandas-dev/pandas... | https://api.github.com/repos/pandas-dev/pandas... | https://github.com/pandas-dev/pandas/issues/38069 | 751083108 | MDU6SXNzdWU3NTEwODMxMDg= | 38069 | ENH: Rename multi-level columns or indices usi... | ... | None | 0 | 2020-11-25 19:57:28+00:00 | 2020-11-25 19:57:28+00:00 | NaT | NONE | NaN | NaN | It's currently quite difficult to rename a sin... | NaN |
| 2 | https://api.github.com/repos/pandas-dev/pandas... | https://api.github.com/repos/pandas-dev/pandas | https://api.github.com/repos/pandas-dev/pandas... | https://api.github.com/repos/pandas-dev/pandas... | https://api.github.com/repos/pandas-dev/pandas... | https://github.com/pandas-dev/pandas/pull/38068 | 751038480 | MDExOlB1bGxSZXF1ZXN0NTI3NjI3Njc2 | 38068 | ENH: NDArrayBackedExtensionArray.__array_funct... | ... | None | 1 | 2020-11-25 18:32:41+00:00 | 2020-11-25 18:49:53+00:00 | NaT | MEMBER | NaN | {'url': 'https://api.github.com/repos/pandas-d... | motivated by getting np.delete and np.repeat w... | NaN |
| 3 | https://api.github.com/repos/pandas-dev/pandas... | https://api.github.com/repos/pandas-dev/pandas | https://api.github.com/repos/pandas-dev/pandas... | https://api.github.com/repos/pandas-dev/pandas... | https://api.github.com/repos/pandas-dev/pandas... | https://github.com/pandas-dev/pandas/issues/38067 | 750998859 | MDU6SXNzdWU3NTA5OTg4NTk= | 38067 | DOC: Include missing holiday observance rules | ... | None | 0 | 2020-11-25 17:25:35+00:00 | 2020-11-25 17:28:13+00:00 | NaT | NONE | NaN | NaN | #### Location of the documentation\r\n\r\n[Tim... | NaN |
| 4 | https://api.github.com/repos/pandas-dev/pandas... | https://api.github.com/repos/pandas-dev/pandas | https://api.github.com/repos/pandas-dev/pandas... | https://api.github.com/repos/pandas-dev/pandas... | https://api.github.com/repos/pandas-dev/pandas... | https://github.com/pandas-dev/pandas/issues/38066 | 750932114 | MDU6SXNzdWU3NTA5MzIxMTQ= | 38066 | RLS: 1.1.5 | ... | {'url': 'https://api.github.com/repos/pandas-d... | 3 | 2020-11-25 15:51:33+00:00 | 2020-11-25 16:11:36+00:00 | NaT | MEMBER | NaN | NaN | Tracking issue for the 1.1.5 release.\r\n\r\nh... | NaN |
5 rows × 26 columns
Navigating a DataFrame
Elements or subsets of a DataFrame can be accessed in multiple ways. We can use [] or use slice notation, marked by the colon (:) character to access subsets of data. Indexing and slicing to select subsets of a DataFrame can be performed:
- By location, like lists
- By label, like dictionaries
- By boolean masks like NumPy arrays
Special indexing operators such as loc and iloc also enable selection of a subset of the rows and columns from a DataFrame. Let's look at some examples.
Selecting Columns
Columns of the DataFrame are essentially Series objects that can be accessed via dictionary-style indexing.
df2['population']CA 39.50 TX 29.00 NY 8.39 FL 21.48 IL 12.67 Name: population, dtype: float64
Individual columns can also be selected using attribute-style access.
df2.populationCA 39.50 TX 29.00 NY 8.39 FL 21.48 IL 12.67 Name: population, dtype: float64
Selection using loc and iloc
Special indexing operators such as loc and iloc can be used to select a subset of the rows and columns from a DataFrame.
.locfor label-based indexing can be used to index the data in an array-like style by specifying index and column names.ilocfor positional indexing can be used to index the underlying array as if it is a simple NumPy array.
Let's add another column to df2 DataFrame and then look at some examples.
# Add column
df2['density'] = df2['population']*1000000 / df2['land area']
df2| population | land area | density | |
|---|---|---|---|
| CA | 39.50 | 155779.22 | 253.563986 |
| TX | 29.00 | 261231.71 | 111.012557 |
| NY | 8.39 | 47126.40 | 178.031846 |
| FL | 21.48 | 53624.76 | 400.561233 |
| IL | 12.67 | 55518.93 | 228.210450 |
Selection using .iloc
.iloc is used for positional indexing.
# First two rows and all columns
df2.iloc[:2, :]| population | land area | density | |
|---|---|---|---|
| CA | 39.5 | 155779.22 | 253.563986 |
| TX | 29.0 | 261231.71 | 111.012557 |
# Last two rows and first two columns
df2.iloc[-2:, :2]| population | land area | |
|---|---|---|
| FL | 21.48 | 53624.76 |
| IL | 12.67 | 55518.93 |
# Middle subset of rows and columns
df2.iloc[2:4, 1:2]| land area | |
|---|---|
| NY | 47126.40 |
| FL | 53624.76 |
Selection using .loc
.loc is used for label-based indexing by specifying index and column names.
# Select rows until 'TX' and all columns
df2.loc[:'TX', :]| population | land area | density | |
|---|---|---|---|
| CA | 39.5 | 155779.22 | 253.563986 |
| TX | 29.0 | 261231.71 | 111.012557 |
# Select rows from 'FL' and columns until 'area'
df2.loc['FL':, :'land area']| population | land area | |
|---|---|---|
| FL | 21.48 | 53624.76 |
| IL | 12.67 | 55518.93 |
# Middle subset of rows from NY to FL and columns from 'area' to 'density'
df2.loc['NY':'FL', 'land area':'density']| land area | density | |
|---|---|---|
| NY | 47126.40 | 178.031846 |
| FL | 53624.76 | 400.561233 |
Conditional Selection
Boolean masks can be used to conditionally select specific subsets of the data. They select the elements of an DataFrame that satisfy some condition. Let's take a look.
df2['density'] > 200CA True TX False NY False FL True IL True Name: density, dtype: bool
# Select data where density > 200
df2[df2['density'] > 200]| population | land area | density | |
|---|---|---|---|
| CA | 39.50 | 155779.22 | 253.563986 |
| FL | 21.48 | 53624.76 | 400.561233 |
| IL | 12.67 | 55518.93 | 228.210450 |
# Select data for only those state where area > 50000 and return first 2 rows
df2[df2['land area'] > 50000].iloc[:2]| population | land area | density | |
|---|---|---|---|
| CA | 39.5 | 155779.22 | 253.563986 |
| TX | 29.0 | 261231.71 | 111.012557 |
Selection using query()
query() enables you to “query” a DataFrame and retrieve subsets based on logical conditions. It acts similar to the ‘where’ clause in SQL making it much easier to read and understand. query() uses string expressions to efficiently compute operations on a DataFrame and offers a more efficient computation compared to the masking expression. It also provides efficient memory use than pure python operations.
Let's add some data to df2 and take a look.
df2['drought'] = ['Yes','Yes','No','Yes','No']df2| population | land area | density | drought | |
|---|---|---|---|---|
| CA | 39.50 | 155779.22 | 253.563986 | Yes |
| TX | 29.00 | 261231.71 | 111.012557 | Yes |
| NY | 8.39 | 47126.40 | 178.031846 | No |
| FL | 21.48 | 53624.76 | 400.561233 | Yes |
| IL | 12.67 | 55518.93 | 228.210450 | No |
Subset based on Numeric Variable
df2.query('population > 20')| population | land area | density | drought | |
|---|---|---|---|---|
| CA | 39.50 | 155779.22 | 253.563986 | Yes |
| TX | 29.00 | 261231.71 | 111.012557 | Yes |
| FL | 21.48 | 53624.76 | 400.561233 | Yes |
Subset based on Categorical Variable
df2.query('drought == "No"')| population | land area | density | drought | |
|---|---|---|---|---|
| NY | 8.39 | 47126.40 | 178.031846 | No |
| IL | 12.67 | 55518.93 | 228.210450 | No |
Subset based on Multiple Conditions
Condition 1: population > 20 and density < 200
# AND operator
df2.query('(population > 20) and (density < 200)')| population | land area | density | drought | |
|---|---|---|---|---|
| TX | 29.0 | 261231.71 | 111.012557 | Yes |
Condition 2: population < 25 or drought == "No"
# OR operator
df2.query('(population < 25) or (drought == "No")')| population | land area | density | drought | |
|---|---|---|---|---|
| NY | 8.39 | 47126.40 | 178.031846 | No |
| FL | 21.48 | 53624.76 | 400.561233 | Yes |
| IL | 12.67 | 55518.93 | 228.210450 | No |
Condition 3: population < 20 and index in ["NY", "IL"]
# IN operator
df2.query('(population < 20) and (index in ["NY", "IL"])')| population | land area | density | drought | |
|---|---|---|---|---|
| NY | 8.39 | 47126.40 | 178.031846 | No |
| IL | 12.67 | 55518.93 | 228.210450 | No |
Condition 4: Like operation
Although "like" is not supported, it can be simulated using string operations and specifying engine='python'.
df2.query('drought.str.contains("e")', engine='python')| population | land area | density | drought | |
|---|---|---|---|---|
| CA | 39.50 | 155779.22 | 253.563986 | Yes |
| TX | 29.00 | 261231.71 | 111.012557 | Yes |
| FL | 21.48 | 53624.76 | 400.561233 | Yes |
Escape special characters
land area column has a whitespace which may cause issues when a query gets executed. Wrap column names in backticks to escape special characters such as whitespace etc.
df2.query('`land area` > 60000')| population | land area | density | drought | |
|---|---|---|---|---|
| CA | 39.5 | 155779.22 | 253.563986 | Yes |
| TX | 29.0 | 261231.71 | 111.012557 | Yes |
Data Types in a DataFrame
The way information is stored in a DataFrame affects what we can do with it and the outputs of calculations on it. When doing data analysis, it is important to make sure you are using the correct data types; otherwise you may get unexpected results or errors. Common data types available in Pandas are object, int64, float64, datetime64 and bool.
An object data type can contain multiple different types such as integers, floats and strings. These different data types when included in a single column are collectively labeled as an object.
Let's expand the df2 DataFrame to add month, year, date, gdp, rainfall and drought columns and explore various data types.
# Add Columns
df2['year'] = [2019,2019,2020,2019,2020]
df2['month'] = [2,5,7,9,11]
df2['day'] = [27,12,10,7,17]
df2['gdp'] = ['3183', '1918', '1751', '1111', '908']
df2['rainfall'] = ['22.9',35,42.87,'54.73','No Value']
df2| population | land area | density | drought | year | month | day | gdp | rainfall | |
|---|---|---|---|---|---|---|---|---|---|
| CA | 39.50 | 155779.22 | 253.563986 | Yes | 2019 | 2 | 27 | 3183 | 22.9 |
| TX | 29.00 | 261231.71 | 111.012557 | Yes | 2019 | 5 | 12 | 1918 | 35 |
| NY | 8.39 | 47126.40 | 178.031846 | No | 2020 | 7 | 10 | 1751 | 42.87 |
| FL | 21.48 | 53624.76 | 400.561233 | Yes | 2019 | 9 | 7 | 1111 | 54.73 |
| IL | 12.67 | 55518.93 | 228.210450 | No | 2020 | 11 | 17 | 908 | No Value |
Checking Data Types
Using the dtypes property of a DataFrame, we can check the different Data Types of each column in a DataFrame.
df2.dtypespopulation float64 land area float64 density float64 drought object year int64 month int64 day int64 gdp object rainfall object dtype: object
info() function can also be used to get a more detailed information; however, it is a more time consuming operation.
df2.info()<class 'pandas.core.frame.DataFrame'> Index: 5 entries, CA to IL Data columns (total 9 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 population 5 non-null float64 1 land area 5 non-null float64 2 density 5 non-null float64 3 drought 5 non-null object 4 year 5 non-null int64 5 month 5 non-null int64 6 day 5 non-null int64 7 gdp 5 non-null object 8 rainfall 5 non-null object dtypes: float64(3), int64(3), object(3) memory usage: 560.0+ bytes
By looking at the data types of different columns, we can identify a few issues:
year,monthanddaycolumns are of typeint64and should be converted todatetime64.gdpis of typeobjectand should befloat64.drought, a Yes/No column is of typeobjectand should be converted tobool.rainfallis also of typeobjectand should be converted tofloat64
Changing Data Types
In Pandas, there are three options to convert the data type of a column:
- Using
astype()function to to force the appropriate data type. - Writing a custom function.
- Using pandas built-in functions such as
to_numeric()andto_datetime().
Let's look at each of these options.
Using astype() function
astype() is the simplest way to convert the data type. Let's start by converting the gdp column of type object to float64 data type.
df2['gdp'].astype('float')CA 3183.0 TX 1918.0 NY 1751.0 FL 1111.0 IL 908.0 Name: gdp, dtype: float64
df2.dtypespopulation float64 land area float64 density float64 drought object year int64 month int64 day int64 gdp object rainfall object dtype: object
Simply running astype() on a column only returns a copy of the column. To ensure the change gets applied to the DataFrame, we need to assign it back.
# Change data type
df2['gdp'] = df2['gdp'].astype('float')
# Check data type
df2['gdp'].dtypedtype('float64')Now let's try changing the data types of drought (dtype object) to bool and rainfall (dtype object) to float64.
Change drought to bool type
df2['drought'].astype('bool')CA True TX True NY True FL True IL True Name: drought, dtype: bool
Here we see that after type conversion, all values are interpreted as
True, however, the column consisted of bothYesandNovalues. By default, if there is a value in column, python evaluates it toTrueirrespective of the type of value. Learn more about truth value evaluation here.
Change rainfall to float64 type

This results in a
ValueErrorwhich means the conversion ofrainfalltofloat64data type did not work.
astype() works when the data is clean and when you want to convert a numeric to a string object. However, when the data is not homogeneous (i.e. object) or has characters, astype may not be the right way to change the data type of a column.
Let's look at the other options of converting data types (mentioned above) to see if we can fix these issues.
Custom Functions
We can write custom functions and then apply them to convert the data type of a column to an appropriate type. Let's change the data type of drought (dtype object) to bool.
# Create function
def str2bool(val):
"""
Converts a string data type to boolean
- converts string values to lowercase
- checks for multiple positive values
- returns the correct boolean type
"""
val = val.lower() # converts string value to lowercase
if val in ("yes", "y", "true", "t", "1"): # checks for multiple positive values
return True
return FalseWe can now use the apply function to apply the str2bool function to all values of the column.
Remember that simply applying a function to a column only returns a copy of the column. To ensure the change gets applied to the DataFrame, we need to assign it back.
# Apply the function
df2['drought'] = df2['drought'].apply(str2bool)
# Check type
df2['drought'].dtypedtype('bool')# Look at column values
df2['drought']CA True TX True NY False FL True IL False Name: drought, dtype: bool
The beauty of custom functions is that they open up a gateway of opportunities. With a simple function, we could consider multiple string values such as "yes", "y", "true", "t", "1". As a result, the column can essentially have any of these values and the function will return True.
This was just a simple function and much more complex functions can be written to address various issues in the data.
Using np.where()
Another popular approach that is useful for many types of problems is to use np.where() function. Let's convert the data type of drought back to object and then take a look at using np.where().
df2['drought'] = ['Y','Y','N','Y','N']
df2['drought'].dtypedtype('O')# Use np.where()
df2['drought'] = np.where(df2['drought'] == 'Y', True, False)# Check values
df2['drought']CA True TX True NY False FL True IL False Name: drought, dtype: bool
np.where() function converts allY values to True and everything else is changed to False.
Pandas built-in functions
Pandas built-in helper functions, such as to_numeric() and to_datetime(), can be very useful for converting certain data type.
Convert rainfall column (type object) to float
# Look at rainfall column
df2['rainfall']CA 22.9 TX 35 NY 42.87 FL 54.73 IL No Value Name: rainfall, dtype: object
# Check the data type of each value in the column
df2['rainfall'].apply(type)CA <class 'str'> TX <class 'int'> NY <class 'float'> FL <class 'str'> IL <class 'str'> Name: rainfall, dtype: object
The rainfall column contains values of multiple different types, such as integers, floats and strings. Let's use the to_numeric function to change data type to float64.
pd.to_numeric(df2['rainfall'], errors='coerce')CA 22.90 TX 35.00 NY 42.87 FL 54.73 IL NaN Name: rainfall, dtype: float64
By passing errors=coerce, the function replaces the invalid “No Value” value with a NaN.
To ensure the change gets applied to the DataFrame, we need to assign it back. Here is another way of applying to_numeric using the apply function.
df2['rainfall'] = df2['rainfall'].apply(pd.to_numeric, errors='coerce')df2['rainfall']CA 22.90 TX 35.00 NY 42.87 FL 54.73 IL NaN Name: rainfall, dtype: float64
Convert year, month, day columns (type int) to datetime64
The year, month, day columns can be combined into a single new date column with the correct data type. Let's convert the date columns to datetime64 type using pd.to_datetime(). We will dive into the details of individual date or time columns in a later section of this guide series.
df2['date'] = pd.to_datetime(df2[['year','month','day']])df2['date']CA 2019-02-27 TX 2019-05-12 NY 2020-07-10 FL 2019-09-07 IL 2020-11-17 Name: date, dtype: datetime64[ns]
Now, the data types of df2 DataFrame are all cleaned up. Let's take a look.
df2.dtypespopulation float64 land area float64 density float64 drought bool year int64 month int64 day int64 gdp float64 rainfall float64 date datetime64[ns] dtype: object
Operating on DataFrames
Pandas is designed to work with NumPy and essentially inherits the ability to perform quick element-wise operations with basic arithmetic (add, subtract, multiply, etc.) and more sophisticated operations (trigonometric, exponential and logarithmic functions, etc.) from NumPy.
There are various other ways in which users can interact with the data in a DataFrame, such as reindexing data, dropping data, adding data, renaming columns etc. Let's explore how we can operate on the data in a DataFrame.
Arithmetic Operations
In Pandas,
- Unary ufuncs (operate on a single input): such as exponential and logarithmic functions, preserve index and column labels in the output.
- Binary ufuncs (operate on two inputs): such as addition and multiplication, automatically align indices and return a DataFrame whose index and columns are the unions of the ones in each DataFrame.
Let's create sample data to demonstrate this.
# Create data
rng = np.random.RandomState(55)
df3 = pd.DataFrame(rng.randint(1,20,(3,3)), columns=list('bcd'),
index=['Ohio', 'Texas', 'Colorado'])
print('df3 is: \n', df3)
print()
df4 = pd.DataFrame(rng.randint(1,25,(4,3)), columns=list('bde'),
index=['Arizona', 'Ohio', 'Texas', 'Iowa'])
print('df4 is: \n', df4)df3 is:
b c d
Ohio 14 8 9
Texas 6 6 2
Colorado 17 4 15
df4 is:
b d e
Arizona 14 13 16
Ohio 8 2 22
Texas 24 9 20
Iowa 11 8 1
Unary Operations
When applying a NumPy ufunc on DataFrame object, the result will be a Pandas object with the indices preserved.
print('Exponentiation:\n', np.power(df3,2))
print()
print('Square Root:\n', np.sqrt(df3))
print()
print("ln(x)\n", np.log(df4))Exponentiation:
b c d
Ohio 196 64 81
Texas 36 36 4
Colorado 289 16 225
Square Root:
b c d
Ohio 3.741657 2.828427 3.000000
Texas 2.449490 2.449490 1.414214
Colorado 4.123106 2.000000 3.872983
ln(x)
b d e
Arizona 2.639057 2.564949 2.772589
Ohio 2.079442 0.693147 3.091042
Texas 3.178054 2.197225 2.995732
Iowa 2.397895 2.079442 0.000000
Binary Operations
Pandas will automatically align indices and return a DataFrame whose index and columns are the unions of the ones in each DataFrame.
# Addition
df3 + df4| b | c | d | e | |
|---|---|---|---|---|
| Arizona | NaN | NaN | NaN | NaN |
| Colorado | NaN | NaN | NaN | NaN |
| Iowa | NaN | NaN | NaN | NaN |
| Ohio | 22.0 | NaN | 11.0 | NaN |
| Texas | 30.0 | NaN | 11.0 | NaN |
Since the c and e columns are not found in both DataFrame objects, they appear as all missing in the result. The same holds for the rows whose labels are not common in both DataFrame.
We can also use Pandas Methods to perform arithmetic operations. Adding these DataFrame will result in NA values in the locations that don’t overlap.
# Addition
df3.add(df4)| b | c | d | e | |
|---|---|---|---|---|
| Arizona | NaN | NaN | NaN | NaN |
| Colorado | NaN | NaN | NaN | NaN |
| Iowa | NaN | NaN | NaN | NaN |
| Ohio | 22.0 | NaN | 11.0 | NaN |
| Texas | 30.0 | NaN | 11.0 | NaN |
# Subtraction
df3 - df4| b | c | d | e | |
|---|---|---|---|---|
| Arizona | NaN | NaN | NaN | NaN |
| Colorado | NaN | NaN | NaN | NaN |
| Iowa | NaN | NaN | NaN | NaN |
| Ohio | 6.0 | NaN | 7.0 | NaN |
| Texas | -18.0 | NaN | -7.0 | NaN |
# Multiplication
df3 * df4| b | c | d | e | |
|---|---|---|---|---|
| Arizona | NaN | NaN | NaN | NaN |
| Colorado | NaN | NaN | NaN | NaN |
| Iowa | NaN | NaN | NaN | NaN |
| Ohio | 112.0 | NaN | 18.0 | NaN |
| Texas | 144.0 | NaN | 18.0 | NaN |
# Division
df3/df4| b | c | d | e | |
|---|---|---|---|---|
| Arizona | NaN | NaN | NaN | NaN |
| Colorado | NaN | NaN | NaN | NaN |
| Iowa | NaN | NaN | NaN | NaN |
| Ohio | 1.75 | NaN | 4.500000 | NaN |
| Texas | 0.25 | NaN | 0.222222 | NaN |
Operations between DataFrame and Series
Operations between a DataFrame and a Series are similar to the operations between a two-dimensional and one-dimensional NumPy array. The index and column alignment is maintained when applying arithmetic operations between a DataFrame and a Series.
As an example, when we subtract a two-dimensional array and one of its rows, the subtraction is performed once for each row based on NumPy's broadcasting rules. Operations between a DataFrame and a Series are similar.
# Get Data
df3| b | c | d | |
|---|---|---|---|
| Ohio | 14 | 8 | 9 |
| Texas | 6 | 6 | 2 |
| Colorado | 17 | 4 | 15 |
# Subtract row for `Ohio` from df3
df3 - df3.iloc[0]| b | c | d | |
|---|---|---|---|
| Ohio | 0 | 0 | 0 |
| Texas | -8 | -2 | -7 |
| Colorado | 3 | -4 | 6 |
The resulting DataFrame shows element values when the row for Ohio gets subtracted from the DataFrame.
For operating on columns, you can use the object methods while specifying the axis keyword.
df3.subtract(df3['c'], axis='columns')| Colorado | Ohio | Texas | b | c | d | |
|---|---|---|---|---|---|---|
| Ohio | NaN | NaN | NaN | NaN | NaN | NaN |
| Texas | NaN | NaN | NaN | NaN | NaN | NaN |
| Colorado | NaN | NaN | NaN | NaN | NaN | NaN |
The resulting DataFrame shows element values when column c gets subtracted from the DataFrame.
Reindexing Data
reindex is an important Pandas method that conforms the data to a new index. reindex, when applied to a DataFrame, can alter either the (row) index, columns, or both. Lets' take a look at reindexing.
# Create data
df5 = pd.DataFrame(np.arange(9).reshape((3, 3)),
index=['a', 'c', 'e'],
columns=['CA', 'TX', 'OH'])
df5| CA | TX | OH | |
|---|---|---|---|
| a | 0 | 1 | 2 |
| c | 3 | 4 | 5 |
| e | 6 | 7 | 8 |
Reindexing Rows
df5 = df5.reindex(['e','d','c','b','a'])
df5| CA | TX | OH | |
|---|---|---|---|
| e | 6.0 | 7.0 | 8.0 |
| d | NaN | NaN | NaN |
| c | 3.0 | 4.0 | 5.0 |
| b | NaN | NaN | NaN |
| a | 0.0 | 1.0 | 2.0 |
The index for this DataFrame is now updated. For the new indices b and d that were added with reindexing, Pandas has automatically generated Nan values.
Reindexing Columns
Columns can be reindexed using the columns keyword.
df5 = df5.reindex(columns=['CA','OH','TX'])
df5| CA | OH | TX | |
|---|---|---|---|
| e | 6.0 | 8.0 | 7.0 |
| d | NaN | NaN | NaN |
| c | 3.0 | 5.0 | 4.0 |
| b | NaN | NaN | NaN |
| a | 0.0 | 2.0 | 1.0 |
Reindexing Rows and Columns
reindex can be used more succinctly by label-indexing with loc.
df5 = df5.loc[['a','b','c','d','e'], ['TX','OH','CA']]
df5| TX | OH | CA | |
|---|---|---|---|
| a | 1.0 | 2.0 | 0.0 |
| b | NaN | NaN | NaN |
| c | 4.0 | 5.0 | 3.0 |
| d | NaN | NaN | NaN |
| e | 7.0 | 8.0 | 6.0 |
Reset Index
reset_index() can be used to reset the index of a DataFrame to a default index. This operations turns the index labels into columns and assigns a default numerical index to the DataFrame. To modify the DataFrame use inplace=True.
reset_index() comes in handy when working with data that has multiple levels of indexing. We will dig into the details of MultiIndex objects in the next part of this guide series. For now, let's take a quick look at how it works.
df5.reset_index()| index | TX | OH | CA | |
|---|---|---|---|---|
| 0 | a | 1.0 | 2.0 | 0.0 |
| 1 | b | NaN | NaN | NaN |
| 2 | c | 4.0 | 5.0 | 3.0 |
| 3 | d | NaN | NaN | NaN |
| 4 | e | 7.0 | 8.0 | 6.0 |
drop parameter can be used to avoid the old index being added as a column.
df5.reset_index(drop=True)| TX | OH | CA | |
|---|---|---|---|
| 0 | 1.0 | 2.0 | 0.0 |
| 1 | NaN | NaN | NaN |
| 2 | 4.0 | 5.0 | 3.0 |
| 3 | NaN | NaN | NaN |
| 4 | 7.0 | 8.0 | 6.0 |
Dropping Data
Dropping rows or columns comes in handy when cleaning your data. The drop() function can be easily applied.
Functions that modify the size or shape of a DataFrame return a new object so that the original data remains unchanged. To apply changes to existing DataFrame, we need to either assign the function back to DataFrame or use inplace keyword.
Dropping rows
Calling drop with index labels will drop values from the row.
# Drop rows
df5.drop(['b','d'])| TX | OH | CA | |
|---|---|---|---|
| a | 1.0 | 2.0 | 0.0 |
| c | 4.0 | 5.0 | 3.0 |
| e | 7.0 | 8.0 | 6.0 |
The result of drop operation is a new object as it does not modify the original DataFrame.
df5| TX | OH | CA | |
|---|---|---|---|
| a | 1.0 | 2.0 | 0.0 |
| b | NaN | NaN | NaN |
| c | 4.0 | 5.0 | 3.0 |
| d | NaN | NaN | NaN |
| e | 7.0 | 8.0 | 6.0 |
To modify the DataFrame, we will use inplace keyword.
df5.drop(['b','d'], inplace=True)
df5| TX | OH | CA | |
|---|---|---|---|
| a | 1.0 | 2.0 | 0.0 |
| c | 4.0 | 5.0 | 3.0 |
| e | 7.0 | 8.0 | 6.0 |
Dropping Columns
Columns can be dropped by passing a value to the axis keyword: axis=1 or axis='columns'.
df5.drop('CA', axis='columns')| TX | OH | |
|---|---|---|
| a | 1.0 | 2.0 |
| c | 4.0 | 5.0 |
| e | 7.0 | 8.0 |
Adding Data
A key aspect of data exploration is to feature engineer the data i.e. add new features to the data. New columns can be easily added to a DataFrame using the following methods.
Using Values in a List
A new column can be added with default values from a list.
df5['FL'] = [5,8,10]
df5| TX | OH | CA | FL | |
|---|---|---|---|---|
| a | 1.0 | 2.0 | 0.0 | 5 |
| c | 4.0 | 5.0 | 3.0 | 8 |
| e | 7.0 | 8.0 | 6.0 | 10 |
The column for FL is now added to the end of the DataFrame.
Using insert()
insert() provides the flexibility to add a column at any index position. It also provides different options for inserting the column values. Let's add a new column for NY at index position 2 between OH and CA.
df5.insert(loc=2, column='NY', value=[7,4,9])
df5| TX | OH | NY | CA | FL | |
|---|---|---|---|---|---|
| a | 1.0 | 2.0 | 7 | 0.0 | 5 |
| c | 4.0 | 5.0 | 4 | 3.0 | 8 |
| e | 7.0 | 8.0 | 9 | 6.0 | 10 |
Using assign()
assign() returns a new DataFrame object that has a copy of the original data with the requested changes. The original DataFrame remains unchanged. To apply changes to existing DataFrame, we need to assign the function back to the DataFrame.
df5 = df5.assign(WA = [0,3,12])df5| TX | OH | NY | CA | FL | WA | |
|---|---|---|---|---|---|---|
| a | 1.0 | 2.0 | 7 | 0.0 | 5 | 0 |
| c | 4.0 | 5.0 | 4 | 3.0 | 8 | 3 |
| e | 7.0 | 8.0 | 9 | 6.0 | 10 | 12 |
Renaming Columns
Real world data is messy. In your data exploration journey, you may come across column names that are not representative of the data or that are too long, or you may just want to standardize the names of columns in your dataset. Let's look at how we can rename columns.
Using rename()
rename() is quite useful when we need to rename some selected columns. Let's rename some columns to reflect the names of states.
df5.rename(columns = {'TX':'Texas', 'OH':'Ohio'}, inplace=True)df5| Texas | Ohio | NY | CA | FL | WA | |
|---|---|---|---|---|---|---|
| a | 1.0 | 2.0 | 7 | 0.0 | 5 | 0 |
| c | 4.0 | 5.0 | 4 | 3.0 | 8 | 3 |
| e | 7.0 | 8.0 | 9 | 6.0 | 10 | 12 |
By assigning a list of Columns
We can also assign a list of new names to the columns attribute of the DataFrame object. The caveat is that this method required providing new names for all the columns even if want to rename only a few.
df5.columns = ['Texas', 'Ohio', 'New York', 'California', 'Florida', 'Washington']df5| Texas | Ohio | New York | California | Florida | Washington | |
|---|---|---|---|---|---|---|
| a | 1.0 | 2.0 | 7 | 0.0 | 5 | 0 |
| c | 4.0 | 5.0 | 4 | 3.0 | 8 | 3 |
| e | 7.0 | 8.0 | 9 | 6.0 | 10 | 12 |
Working with Missing Data
Missing data occurs in many applications as real world data in rarely clean. It is generally referred to as Null, NaN, or NA values. Assessing the missing data is an important process, as it can identify potential problems or biases that may arise as a result of the missing data. There are various useful methods for detecting, removing, and replacing null values in Pandas such as:
isnull(): generates a boolean mask indicating missing values.notnull(): generates a boolean mask indicating non-missing values.dropna(): drops null values and returns a filtered version of the data.fillna(): returns a copy of the data with missing values filled or imputed.interpolate(): powerful function that providers various interpolation techniques to fill the missing values.
Let's start by looking at the types of missing data in Pandas and then we will explore how to detect, filter, drop and impute missing data.
Missing Data in Pandas
Pandas uses two already existing Python null values:
None- is a Python singleton object only used in arrays with data type 'object'.NaN- is a special floating point value.
None object
v1 = np.array([1, None, 4, 6])
v1array([1, None, 4, 6], dtype=object)
dtype=object shows NumPy inferred that the contents of this array are Python objects. Any operations on the data will be done at the Python level, which are typically slower than the arrays with native types.
for dtype in ['object', 'int', 'float']:
print("dtype =", dtype)
%timeit np.arange(1E4, dtype=dtype).sum()
print()dtype = object 391 µs ± 5.76 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each) dtype = int 11.2 µs ± 708 ns per loop (mean ± std. dev. of 7 runs, 100000 loops each) dtype = float 11.2 µs ± 653 ns per loop (mean ± std. dev. of 7 runs, 100000 loops each)
Aggregation operations on an array with None value results in an error.

NaN: Not a Number
v2 = np.array([1, np.NaN, 4, 6])
v2.dtypedtype('float64')dtype('float64') shows NumPy inferred that the contents of this array are native floating-point type. Unlike object, this array supports faster operations.
Aggregation operations on an array with NaN will result in a NaN.
v2.sum(), v2.mean()(nan, nan)
NaN and None
Pandas handles both NaN and None interchangeably and automatically converts them as appropriate.
pd.Series([1, np.nan, 5, None])0 1.0 1 NaN 2 5.0 3 NaN dtype: float64
Pandas automatically converts None to a NaN value.
Detect Missing Data
isnull() and notnull() methods can be used to detect and filter missing data. We will start by creating some data to apply these methods.
# Create data
df6 = df3 + df4
df6.loc['Ohio','c'] = 10
df6.loc['Colorado','b'] = 5
df6| b | c | d | e | |
|---|---|---|---|---|
| Arizona | NaN | NaN | NaN | NaN |
| Colorado | 5.0 | NaN | NaN | NaN |
| Iowa | NaN | NaN | NaN | NaN |
| Ohio | 22.0 | 10.0 | 11.0 | NaN |
| Texas | 30.0 | NaN | 11.0 | NaN |
df6.isnull()| b | c | d | e | |
|---|---|---|---|---|
| Arizona | True | True | True | True |
| Colorado | False | True | True | True |
| Iowa | True | True | True | True |
| Ohio | False | False | False | True |
| Texas | False | True | False | True |
The result shows a matrix of True or False values based on the presence of null values in data.
Missing values in each column
Chaining a sum() method returns a summation of missing values in each column.
df6.isnull().sum()b 2 c 4 d 3 e 5 dtype: int64
Missing values in DataFrame
Chaining two sum() methods will return the total number of missing values in the DataFrame.
df6.isnull().sum().sum()14
Using notnull()
notnull() is the opposite of isnull() and can be used to check the number of non-missing values.
df6.notnull().sum()b 3 c 1 d 2 e 0 dtype: int64
# Total non-missing values in dataframe
df6.notnull().sum().sum()6
Visualize Missing Data
It is important to understand the pattern of occurrence of missing values before we decide to drop or impute missing data. An easy way to visualize missing records is to use heatmap() from the seaborn library.
# Import Seaborn
import seaborn as sns# Get Data
df6| b | c | d | e | |
|---|---|---|---|---|
| Arizona | NaN | NaN | NaN | NaN |
| Colorado | 5.0 | NaN | NaN | NaN |
| Iowa | NaN | NaN | NaN | NaN |
| Ohio | 22.0 | 10.0 | 11.0 | NaN |
| Texas | 30.0 | NaN | 11.0 | NaN |
# Visualize missing
sns.heatmap(df6.isnull(), cbar=False, cmap='viridis')<matplotlib.axes._subplots.AxesSubplot at 0x7fb2e87b5a90>
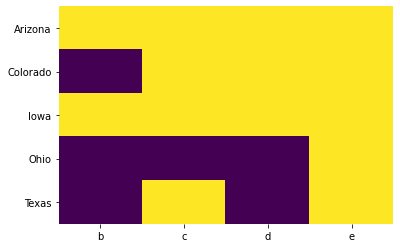
Missing records are displayed in yellow color.
Filter based on Missing Data
Both isnull() and notnull() can be applied to columns of a DataFrame to filter out rows with missing and non-missing data.
# Get data
df6| b | c | d | e | |
|---|---|---|---|---|
| Arizona | NaN | NaN | NaN | NaN |
| Colorado | 5.0 | NaN | NaN | NaN |
| Iowa | NaN | NaN | NaN | NaN |
| Ohio | 22.0 | 10.0 | 11.0 | NaN |
| Texas | 30.0 | NaN | 11.0 | NaN |
Filter rows where values in column b are null.
df6[df6['b'].isnull()]| b | c | d | e | |
|---|---|---|---|---|
| Arizona | NaN | NaN | NaN | NaN |
| Iowa | NaN | NaN | NaN | NaN |
Filter rows where values in column b are not null.
df6[df6['b'].notnull()]| b | c | d | e | |
|---|---|---|---|---|
| Colorado | 5.0 | NaN | NaN | NaN |
| Ohio | 22.0 | 10.0 | 11.0 | NaN |
| Texas | 30.0 | NaN | 11.0 | NaN |
Drop Missing Data
dropna() method allows us to drop any missing values from the data. The method returns a copy of the DataFrame, so to apply the changes inplace, use inplace=True.
# Get data
df6| b | c | d | e | |
|---|---|---|---|---|
| Arizona | NaN | NaN | NaN | NaN |
| Colorado | 5.0 | NaN | NaN | NaN |
| Iowa | NaN | NaN | NaN | NaN |
| Ohio | 22.0 | 10.0 | 11.0 | NaN |
| Texas | 30.0 | NaN | 11.0 | NaN |
By default, dropna() will drop all rows in which any null value is present.
df6.dropna()| b | c | d | e |
|---|
Since all rows of df6 have some NA values, the result is an empty copy of the DataFrame. Alternatively, using axis='columns' drops all columns containing a null value.
df6.dropna(axis='columns')| Arizona |
|---|
| Colorado |
| Iowa |
| Ohio |
| Texas |
Since all columns have some NA values, the result is an empty copy of the DataFrame.
dropna() provides the flexibility to drop rows or columns with all NA values, or a majority of NA values using how or thresh parameters. The default how='any', allows any row or column containing a null value to be dropped. To only drop rows or columns that have all null values, how='all' can be specified.
# Drop rows with all null values
df6.dropna(how='all')| b | c | d | e | |
|---|---|---|---|---|
| Colorado | 5.0 | NaN | NaN | NaN |
| Ohio | 22.0 | 10.0 | 11.0 | NaN |
| Texas | 30.0 | NaN | 11.0 | NaN |
# Drop columns with all null values
df6.dropna(how='all', axis='columns')| b | c | d | |
|---|---|---|---|
| Arizona | NaN | NaN | NaN |
| Colorado | 5.0 | NaN | NaN |
| Iowa | NaN | NaN | NaN |
| Ohio | 22.0 | 10.0 | 11.0 |
| Texas | 30.0 | NaN | 11.0 |
thresh parameter allows you specify a minimum number of non-null values a row/column should have to be kept in the result.
df6.dropna(thresh=2)| b | c | d | e | |
|---|---|---|---|---|
| Ohio | 22.0 | 10.0 | 11.0 | NaN |
| Texas | 30.0 | NaN | 11.0 | NaN |
Only rows with 2 or more non-null values are kept, and since the row for Colorado has only 1 non-null value, it is dropped.
df6.dropna(thresh=2, axis='columns')| b | d | |
|---|---|---|
| Arizona | NaN | NaN |
| Colorado | 5.0 | NaN |
| Iowa | NaN | NaN |
| Ohio | 22.0 | 11.0 |
| Texas | 30.0 | 11.0 |
Similarly, column c has only 1 non-null value and is therefore dropped.
Impute Missing Data
Rather than dropping NA values and potentially discarding some other data with it, you may just want to replace them with a value such as 0, or some other imputation such as mean or median of the data. Pandas fillna() method can be used for such operations. The method returns a new object, but you can modify the existing object in-place.
# Get data
df6| b | c | d | e | |
|---|---|---|---|---|
| Arizona | NaN | NaN | NaN | NaN |
| Colorado | 5.0 | NaN | NaN | NaN |
| Iowa | NaN | NaN | NaN | NaN |
| Ohio | 22.0 | 10.0 | 11.0 | NaN |
| Texas | 30.0 | NaN | 11.0 | NaN |
Fill with a Constant Value
Calling fillna() with a constant value replaces missing values with that value.
df6.fillna(0)| b | c | d | e | |
|---|---|---|---|---|
| Arizona | 0.0 | 0.0 | 0.0 | 0.0 |
| Colorado | 5.0 | 0.0 | 0.0 | 0.0 |
| Iowa | 0.0 | 0.0 | 0.0 | 0.0 |
| Ohio | 22.0 | 10.0 | 11.0 | 0.0 |
| Texas | 30.0 | 0.0 | 11.0 | 0.0 |
Forawrd and Backward Fill
Forward and backward fill can be used to propagate the previous value forward (ffill) or next values backward (bfill). We can specify an axis along which the fill method will operate.
Note: If a previous value is not available during a fill operation, the NA value remains.
# Forward fill along rows
df6.fillna(method='ffill')| b | c | d | e | |
|---|---|---|---|---|
| Arizona | NaN | NaN | NaN | NaN |
| Colorado | 5.0 | NaN | NaN | NaN |
| Iowa | 5.0 | NaN | NaN | NaN |
| Ohio | 22.0 | 10.0 | 11.0 | NaN |
| Texas | 30.0 | 10.0 | 11.0 | NaN |
# Forward fill along columns
df6.fillna(method='ffill', axis='columns')| b | c | d | e | |
|---|---|---|---|---|
| Arizona | NaN | NaN | NaN | NaN |
| Colorado | 5.0 | 5.0 | 5.0 | 5.0 |
| Iowa | NaN | NaN | NaN | NaN |
| Ohio | 22.0 | 10.0 | 11.0 | 11.0 |
| Texas | 30.0 | 30.0 | 11.0 | 11.0 |
# Backward fill along rows
df6.fillna(method='bfill')| b | c | d | e | |
|---|---|---|---|---|
| Arizona | 5.0 | 10.0 | 11.0 | NaN |
| Colorado | 5.0 | 10.0 | 11.0 | NaN |
| Iowa | 22.0 | 10.0 | 11.0 | NaN |
| Ohio | 22.0 | 10.0 | 11.0 | NaN |
| Texas | 30.0 | NaN | 11.0 | NaN |
Fill with Arithmetic or Aggregate Functions
A mean, median, mode, max or min value for the column can be used to fill missing values.
# Get data
df6| b | c | d | e | |
|---|---|---|---|---|
| Arizona | NaN | NaN | NaN | NaN |
| Colorado | 5.0 | NaN | NaN | NaN |
| Iowa | NaN | NaN | NaN | NaN |
| Ohio | 22.0 | 10.0 | 11.0 | NaN |
| Texas | 30.0 | NaN | 11.0 | NaN |
# Fill with mean
df6.fillna(df6.mean())| b | c | d | e | |
|---|---|---|---|---|
| Arizona | 19.0 | 10.0 | 11.0 | NaN |
| Colorado | 5.0 | 10.0 | 11.0 | NaN |
| Iowa | 19.0 | 10.0 | 11.0 | NaN |
| Ohio | 22.0 | 10.0 | 11.0 | NaN |
| Texas | 30.0 | 10.0 | 11.0 | NaN |
# Fill with median
df6.fillna(df6.median())/Users/mohi9282/opt/anaconda3/envs/arcgis-clone/lib/python3.8/site-packages/numpy/lib/nanfunctions.py:1115: RuntimeWarning: All-NaN slice encountered r, k = function_base._ureduce(a, func=_nanmedian, axis=axis, out=out,
| b | c | d | e | |
|---|---|---|---|---|
| Arizona | 22.0 | 10.0 | 11.0 | NaN |
| Colorado | 5.0 | 10.0 | 11.0 | NaN |
| Iowa | 22.0 | 10.0 | 11.0 | NaN |
| Ohio | 22.0 | 10.0 | 11.0 | NaN |
| Texas | 30.0 | 10.0 | 11.0 | NaN |
# Fill with max
df6.fillna(df6.max())| b | c | d | e | |
|---|---|---|---|---|
| Arizona | 30.0 | 10.0 | 11.0 | NaN |
| Colorado | 5.0 | 10.0 | 11.0 | NaN |
| Iowa | 30.0 | 10.0 | 11.0 | NaN |
| Ohio | 22.0 | 10.0 | 11.0 | NaN |
| Texas | 30.0 | 10.0 | 11.0 | NaN |
Fill using a Dictionary of Values
A dictionary of constant values or aggregate functions can be passed to fill missing values in columns differently.
df6| b | c | d | e | |
|---|---|---|---|---|
| Arizona | NaN | NaN | NaN | NaN |
| Colorado | 5.0 | NaN | NaN | NaN |
| Iowa | NaN | NaN | NaN | NaN |
| Ohio | 22.0 | 10.0 | 11.0 | NaN |
| Texas | 30.0 | NaN | 11.0 | NaN |
fill_value = {'b': df6['b'].median(),
'd': df6['d'].mean(),
'e': 1}
df6.fillna(value=fill_value)| b | c | d | e | |
|---|---|---|---|---|
| Arizona | 22.0 | NaN | 11.0 | 1.0 |
| Colorado | 5.0 | NaN | 11.0 | 1.0 |
| Iowa | 22.0 | NaN | 11.0 | 1.0 |
| Ohio | 22.0 | 10.0 | 11.0 | 1.0 |
| Texas | 30.0 | NaN | 11.0 | 1.0 |
Using interpolate()
interpolate() is a very powerful function that providers various interpolation techniques (linear, quadratic, polynomial etc.) to fill the missing values. Let's take a quick look, and you can learn more about interpolate() here.
# Get data
df6| b | c | d | e | |
|---|---|---|---|---|
| Arizona | NaN | NaN | NaN | NaN |
| Colorado | 5.0 | NaN | NaN | NaN |
| Iowa | NaN | NaN | NaN | NaN |
| Ohio | 22.0 | 10.0 | 11.0 | NaN |
| Texas | 30.0 | NaN | 11.0 | NaN |
interpolate() using Linear method. Note that Linear method ignores the index and treats the values as equally spaced.
df6.interpolate(method='linear', limit_direction='forward')| b | c | d | e | |
|---|---|---|---|---|
| Arizona | NaN | NaN | NaN | NaN |
| Colorado | 5.0 | NaN | NaN | NaN |
| Iowa | 13.5 | NaN | NaN | NaN |
| Ohio | 22.0 | 10.0 | 11.0 | NaN |
| Texas | 30.0 | 10.0 | 11.0 | NaN |
Conclusion
In this part of the guide series we introduced Pandas, a Python package that builds on NumPy and provides data structures and functions designed to make working with structured data fast, easy, and expressive.
You have seen how DataFrame can be created and then data can be accessed using loc and iloc operators. We discussed in detail how to check the different data types in a DataFrame and ways to change these data types. We also discussed how to perform various operations on a DataFrame (i.e. Arithmetic, Reindex, Add and Drop data) and to work with missing data. We briefly introduced working with a Series object as well.
In the next part of this guide series, you will learn about how to be more productive with Pandas. We will discuss data aggregation, transformation operations, multi-level indexing and working with time series data.
References
[1] Wes McKinney. 2017. Python for Data Analysis: Data Wrangling with Pandas, NumPy, and IPython (2nd. ed.). O'Reilly Media, Inc.
[2] Jake VanderPlas. 2016. Python Data Science Handbook: Essential Tools for Working with Data (1st. ed.). O'Reilly Media, Inc.